The cutlow command¶
The organelle assembler’s cutlow redoes the cleaning step allowing to clean-out all the branches of the assembling graph not reaching the coverage specified by the –coverage option. realizes the assembling of the reads by building the De Bruijn Graph which is the central data structure used by the organelle assembler.
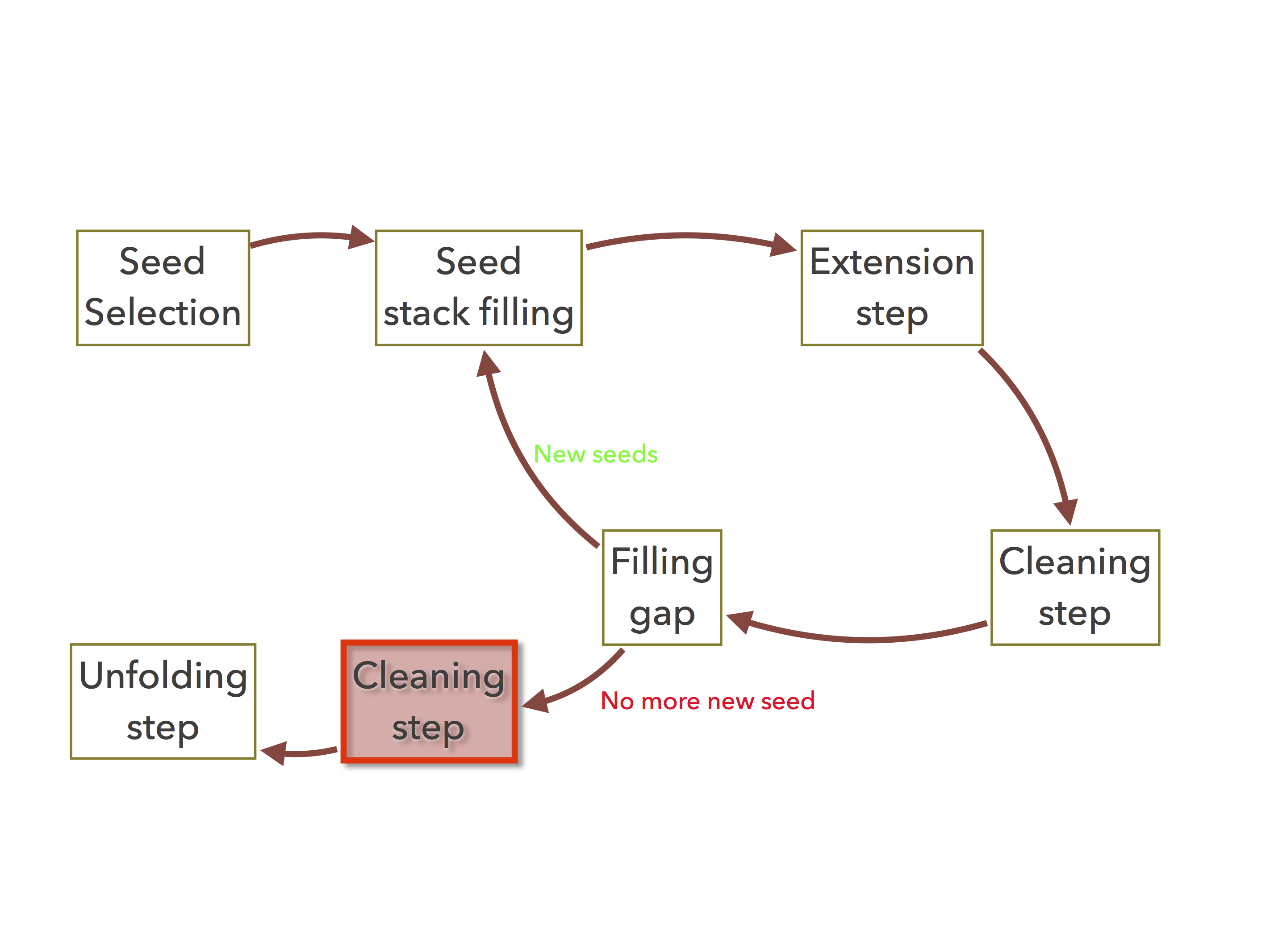
Figure 1: The organelle assembler’s cutlow command re-executes all the cleaning task in red.¶
command prototype¶
usage: oa cutlow [-h]
[--coverage BUILDGRAPH:COVERAGE]
[--smallbranches BUILDGRAPH:SMALLBRANCHES]
[--back ORGASM:BACK] [--snp]
index [output]
positional arguments¶
-
index¶ index root filename (produced by the oa index command)
-
output¶ output prefix
optional arguments¶
Graph cleaning options¶
-
--coverageBUILDGRAPH:COVERAGE¶ during the cutlow execution all stems with a coverage below the specified coverage will be deleted [default:<estimated>]
-
--smallbranchesBUILDGRAPH:SMALLBRANCHES¶ After a cycle a extension, if you observe the assembling graph you can observe a main path and many small aborted branches surrounding this main path. They correspond to path initiated by a sequencing error or a nuclear copy of a chloroplast region not enough covered by the skimming sequencing to be successfully extended. One of the cleaning step consist in deleting these small branches. This option indicates up to which length branches have to be deleted. By default this length is automatically estimated from the graph.
$ oa cutlow --smallbranches 15 seqindexDuring the cleaning steps, all the branches with a length shorter or equal to 15 base pairs will be deleted