The buildgraph command¶
The organelle assembler’s buildgraph realizes the assembling of the reads by building the De Bruijn Graph which is the central data structure used by the organelle assembler.
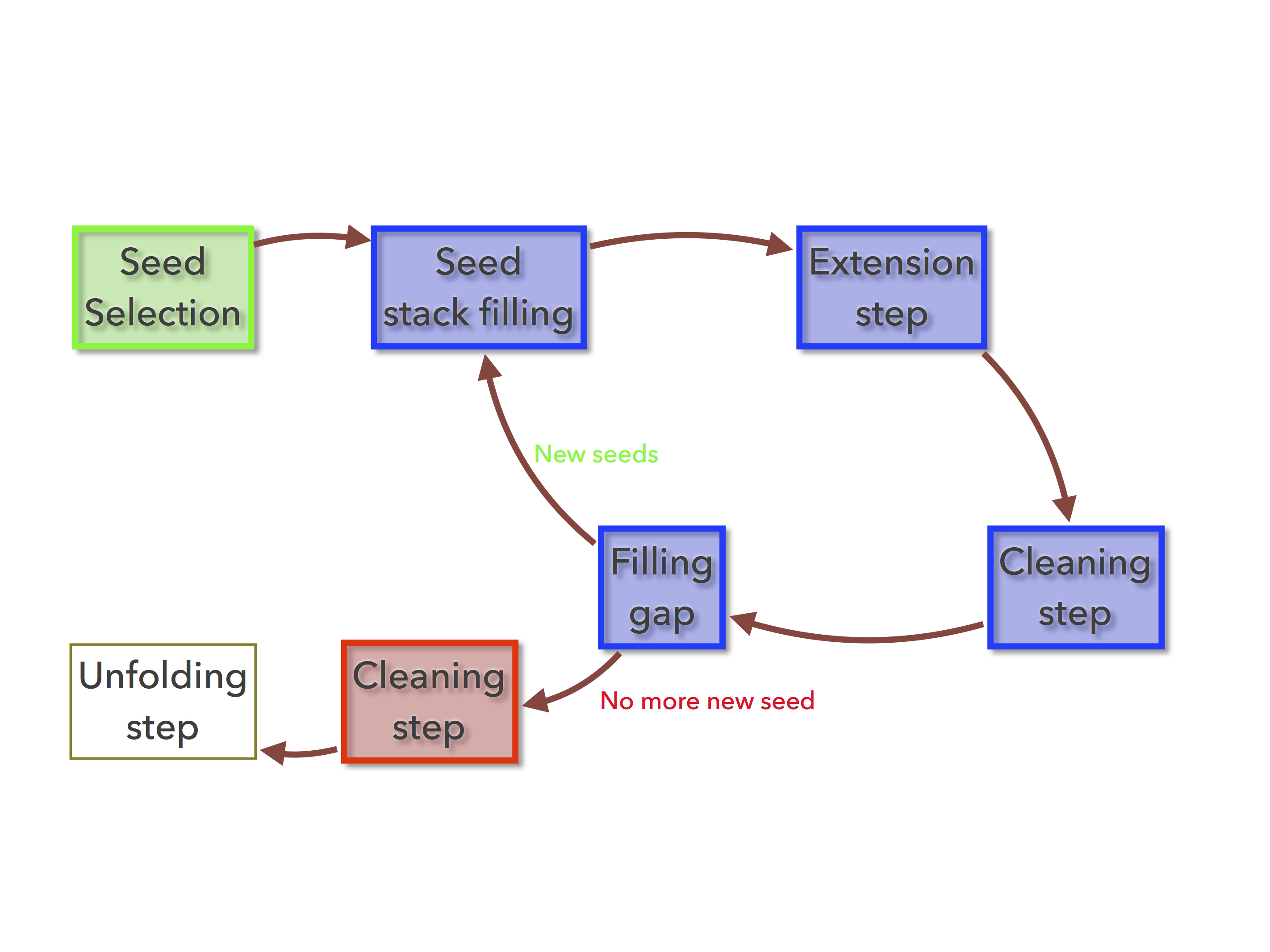
Figure 1: The organelle assembler’s buildgraph command executes all the colored tasks, starting by the green one and ending at the red task¶
command prototype¶
usage: oa buildgraph [-h]
[--reformat]
[--probes probes] [--kup ORGASM:KUP]
[--adapt5 adapt5] [--adapt3 adapt3]
[--phiX] [--phiX-off]
[--coverage BUILDGRAPH:COVERAGE]
[--coverage-ratio BUILDGRAPH:COVERAGE]
[--fillgaps-ratio BUILDGRAPH:COVERAGE]
[--lowcomplexity]
[--minread BUILDGRAPH:MINREAD]
[--minoverlap BUILDGRAPH:MINOVERLAP]
[--minratio BUILDGRAPH:MINRATIO]
[--mincov BUILDGRAPH:MINCOV]
[--assmax BUILDGRAPH:ASSMAX]
[--smallbranches BUILDGRAPH:SMALLBRANCHES]
[--maxfillgaps]
[--clean] [--force-seeds] [--no-seeds seeds]
[--back ORGASM:BACK] [--snp]
index [output]
positional arguments¶
-
index¶ index root filename (produced by the oa index command)
-
output¶ output prefix
optional arguments¶
General option¶
-
-h,--help¶ show the help message and exit
-
--reformat¶ Asks for reformatting an old sequence assembly to the new format
Graph initialisation options¶
-
--seedsseeds¶ Seed sequences; either a fasta file containing seeds sequences (nucleic or proteic) or the name of an internal set of seeds among:
nucrRNAAHypogastruranucrRNAArabidopsisprotChloroArabidopsisprotMitoCapraprotMitoMachaon
$ oa buildgraph --probes protChloroArabidopsis seqindex
A set of seed sequences must be or nucleic or proteic. For initiating
assembling with both nucleic and proteic sequences you must use at least two
--seeds options one for each class of sequences.
$ oa buildgraph --seeds protChloroArabidopsis --seeds rDNAChloro.fasta seqindex
-
--kupORGASM:KUP¶ The word size used to identify the seed reads [default: protein=4, DNA=12].
Graph extension options¶
The main aim of the buildgraph command is to build the De Bruijn Graph which is the central data structure used by the organelle assembler. This building is done by two algorithms:
the extension algorithm which is the main one
the fillgap algorithm which is run when the first one failed to rescue the assembling procedure.
The extension algorithm is actually an heuristics and several parametters can be set to adapt the efficiency of the algorithm to your data. Without precising them, these parameters are automatically estimated from the dataset.
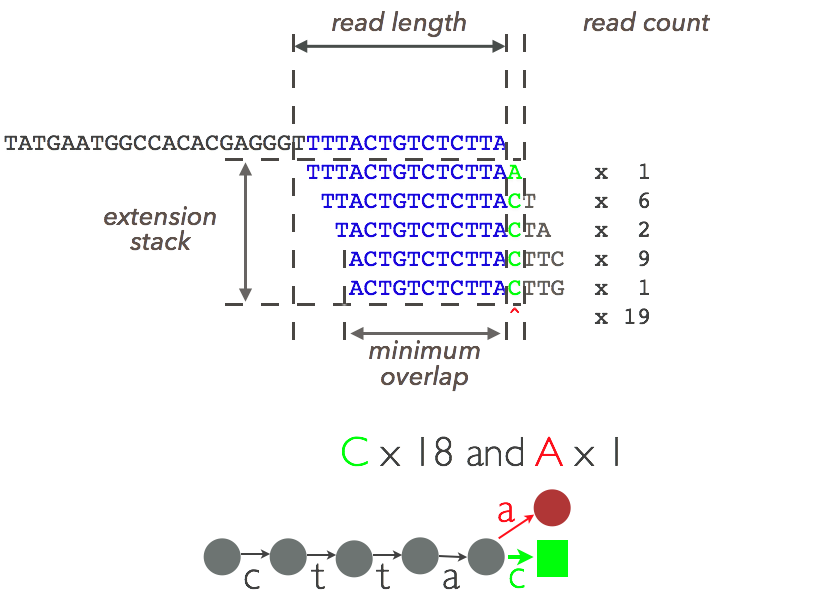
Figure 2: The assembling stack¶
-
--minreadBUILDGRAPH:MINREAD¶ the minimum count of read to consider [default: <estimated>]
$ oa buildgraph --seeds protChloroArabidopsis --minread 5 seqindexConsider an extension if at least five reads are present in the extension stack.
-
--coverageBUILDGRAPH:COVERAGE¶ the expected sequencing coverage [default: <estimated>]
-
--minoverlapBUILDGRAPH:MINOVERLAP¶ minimum length of the overlap between the sequence and reads to participate in the extension. [default: <estimated>]
-
--minratioBUILDGRAPH:MINRATIO¶ minimum ratio between occurrences of an extension and the occurrences of the most frequent extension to keep it. [default: <estimated>]
-
--mincovBUILDGRAPH:MINCOV¶ minimum occurrences of an extension to keep it. [default: 1]
Graph filtering options¶
This set of options allows for excluding some reads from the assembling procedure.
-
--lowcomplexity¶ Use also low complexity probes. Probes are the 3’ end of the sequence currently extended. By default probes with a low complexity are not used during the graph extension procedure. A probe is defined as a low complexity probe if it is fully composed of an homopolymer or an homo-dimer or an homo-trimer.
-
--adapt5adapt5¶ adapter sequences used to filter reads beginning by such sequences; either a fasta file containing adapter sequences or internal set of adapter sequences among [‘adapt5ILLUMINA’] [default: adapt5ILLUMINA]
-
--adapt3adapt3¶ adapter sequences used to filter reads ending by such sequences; either a fasta file containing adapter sequences or internal set of adapter sequences among [‘adapt3ILLUMINA’] [default: adapt3ILLUMINA]
Graph limit option¶
-
--assmaxBUILDGRAPH:ASSMAX¶ Maximum base pair assembled. This limit the size of the De Bruijn Graph, and must be set to a larger value than the size of the sequence you want to assemble to take into account all the alternative paths present in the De Bruijn Graph.
Graph cleaning options¶
-
--smallbranchesBUILDGRAPH:SMALLBRANCHES¶ After a cycle a extension, if you observe the assembling graph you can observe a main path and many small aborted branches surrounding this main path. They correspond to path initiated by a sequencing error or a nuclear copy of a chloroplast region not enough covered by the skimming sequencing to be successfully extended. One of the cleaning step consist in deleting these small branches. This option indicates up to which length branches have to be deleted. By default this length is automatically estimated from the graph.
$ oa buildgraph --seeds protChloroArabidopsis \ --smallbranches 15 seqindex
During the cleaning steps, all the branches with a length shorter or equal to 15 base pairs will be deleted
-
--snp¶ When the data set correspond to a pool of individuals, it is possible that natural polymorphisms artificially complexy the assembling graph. For helping the assembling process of such data set, this option will clear the graph for such SNP by keeping only the most abundant allele prsent in the dataset. The generated sequence can be considered as a king of consensus. Read can be remapped in a second time on this consensus using classical sofware like BWA to get the lost SNP information.
By default this option is deactivated
$ oa buildgraph --seeds protChloroArabidopsis \ --snp seqindexRun the assembling, ignoring the SNPs.