The seeds command¶
Note
For most of the users this command is useless, because this task is automaticaly realized by the oa buildgraph command.
The organelle assembler’s seeds computes the set of seed reads. The main reason of this command if to write a new version of the file containing the set of seed reads, because its format changed.
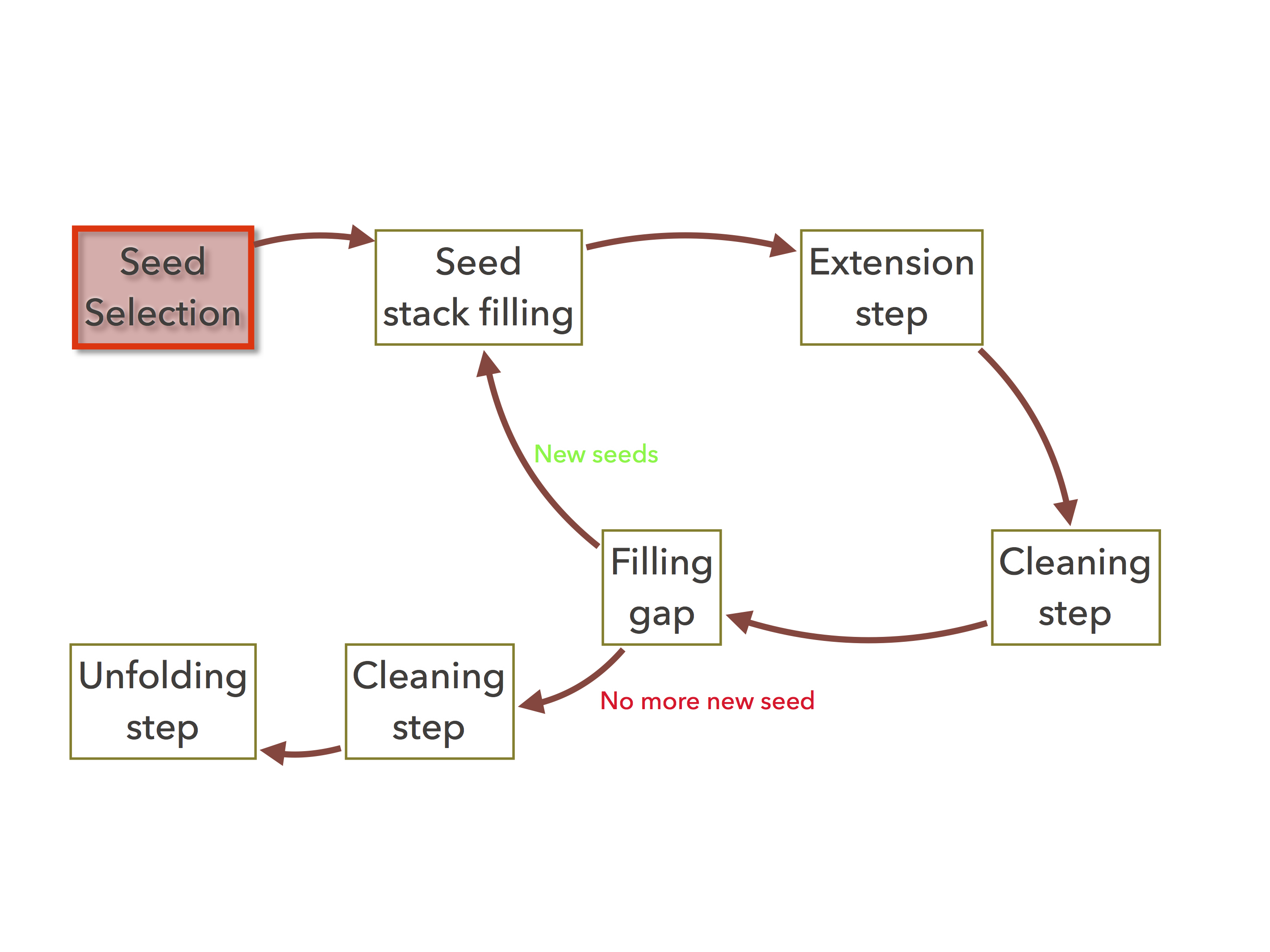
Figure 1: The organelle assembler’s seeds command executes only the red task¶
command prototype¶
usage: oa seeds [-h] [--seeds seeds] [--kup ORGASM:KUP]
index [output]
positional arguments¶
-
index¶ index root filename (produced by the oa index command)
-
output¶ output prefix
optional arguments¶
Graph initialisation options¶
-
--seedsseeds¶ Seed sequences; either a fasta file containing seeds sequences (nucleic or proteic) or the name of an internal set of seeds among:
nucrRNAAHypogastruranucrRNAArabidopsisprotChloroArabidopsisprotMitoCapraprotMitoMachaon
$ oa seeds --seeds protChloroArabidopsis seqindex
A set of seed sequences must be or nucleic or proteic. For initiating
assembling with both nucleic and proteic sequences you must use at least two
--seeds options one for each class of sequences.
$ oa seeds --seeds protChloroArabidopsis --seeds rDNAChloro.fasta seqindex
-
--kupORGASM:KUP¶ The word size used to identify the seed reads [default: protein=4, DNA=12].